Database
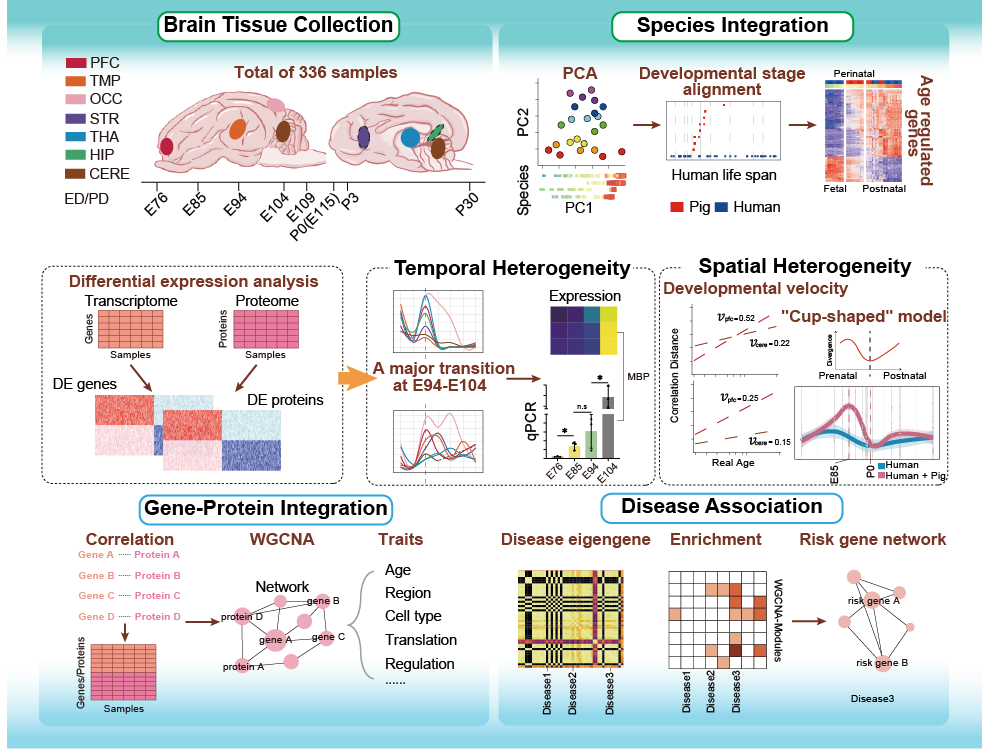
PBAtlas Perinatal Multiome
The perinatal period witnesses dynamic structural reorganization and functional specialization essential for establishing later-life neural circuitry integrity. However, molecular mechanisms governing spatiotemporal developmental trajectories during this critical window remain poorly characterized due to limited accessibility of human perinatal specimens. Here, we analysed transcriptomes and proteomes from 336 samples across multiple time points and brain regions in perinatal Bama miniature pigs, showing their spatiotemporal organization and concordance between genes and proteins. We characterized pigs as optimal large-animal model, demonstrating its closer analogy to humans, resemblance to monkeys, and superiority over the mice and other species, while identifying their perinatal stage as a transitional hub bridging early prenatal to postnatal molecular trajectories in humans. We revealed a pan-regional developmental inflection point (E94-E104) during the perinatal period, manifesting as order-of-magnitude changes in the expression of genes and proteins associated with synaptogenesis and gliogenesis. While further analysing developmental velocities across pig brain regions, we discovered regional heterogeneity exhibits a remarkably synchronized rise-and-fall pattern during the perinatal period. Additionally, brain disorder risk genes exhibited a perinatal transition to an adult-like organization in pigs, establishing this multi-omics atlas as a translational framework for decoding neurodevelopmental disorder pathogenesis.
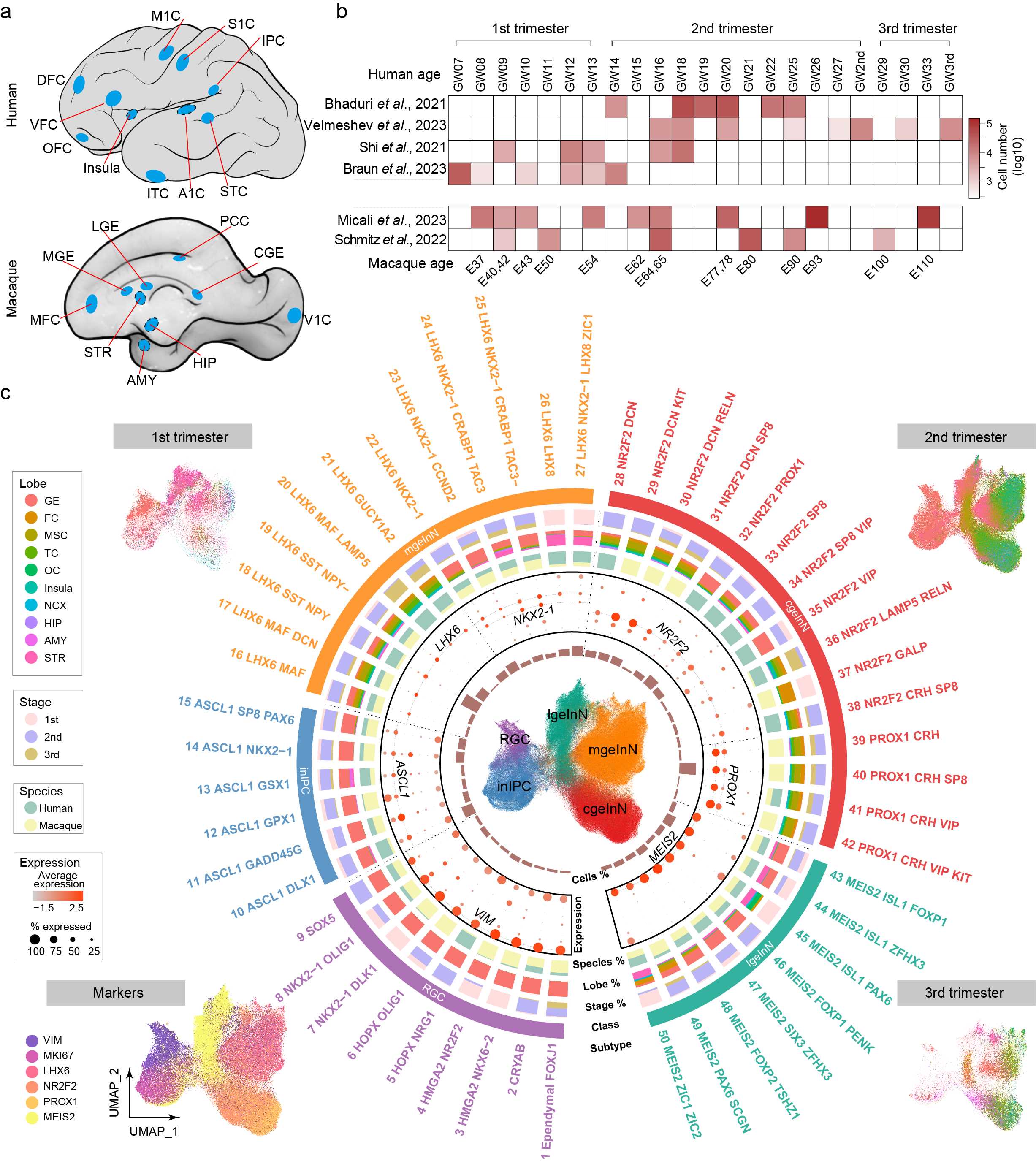
Mammal InN migration
Cortical interneurons generated from ganglionic eminence via a long-distance journey of tangential migration display evident cellular and molecular differences across brain regions, which seeds the heterogeneous cortical circuitry in primates. However, whether such regional specifications in interneurons are intrinsically encoded or gained through interactions with the local milieu remains elusive. Here, we recruit 685,692 interneurons from cerebral cortex and subcortex including ganglionic eminence within the developing human and macaque species. Our integrative and comparative analyses reveal that less transcriptomic alteration is accompanied by interneuron migration within the ganglionic eminence subdivisions, in contrast to the dramatic changes observed in cortical tangential migration, which mostly characterize the transcriptomic specification for different destinations and for species divergence. Moreover, the in-depth survey of temporal regulation illustrates species differences in the developmental dynamics of cell types, e.g., the employment of CRH in primate interneurons during late-fetal stage distinguishes from their postnatal emergence in mice, and our entropy quantifications manifest the interneuron diversities gradually increase along the developmental ages in human and macaque cerebral cortices. Overall, our analyses depict the spatiotemporal features appended to cortical interneurons, providing a new proxy for understanding the relationship between cellular diversity and functional progression.
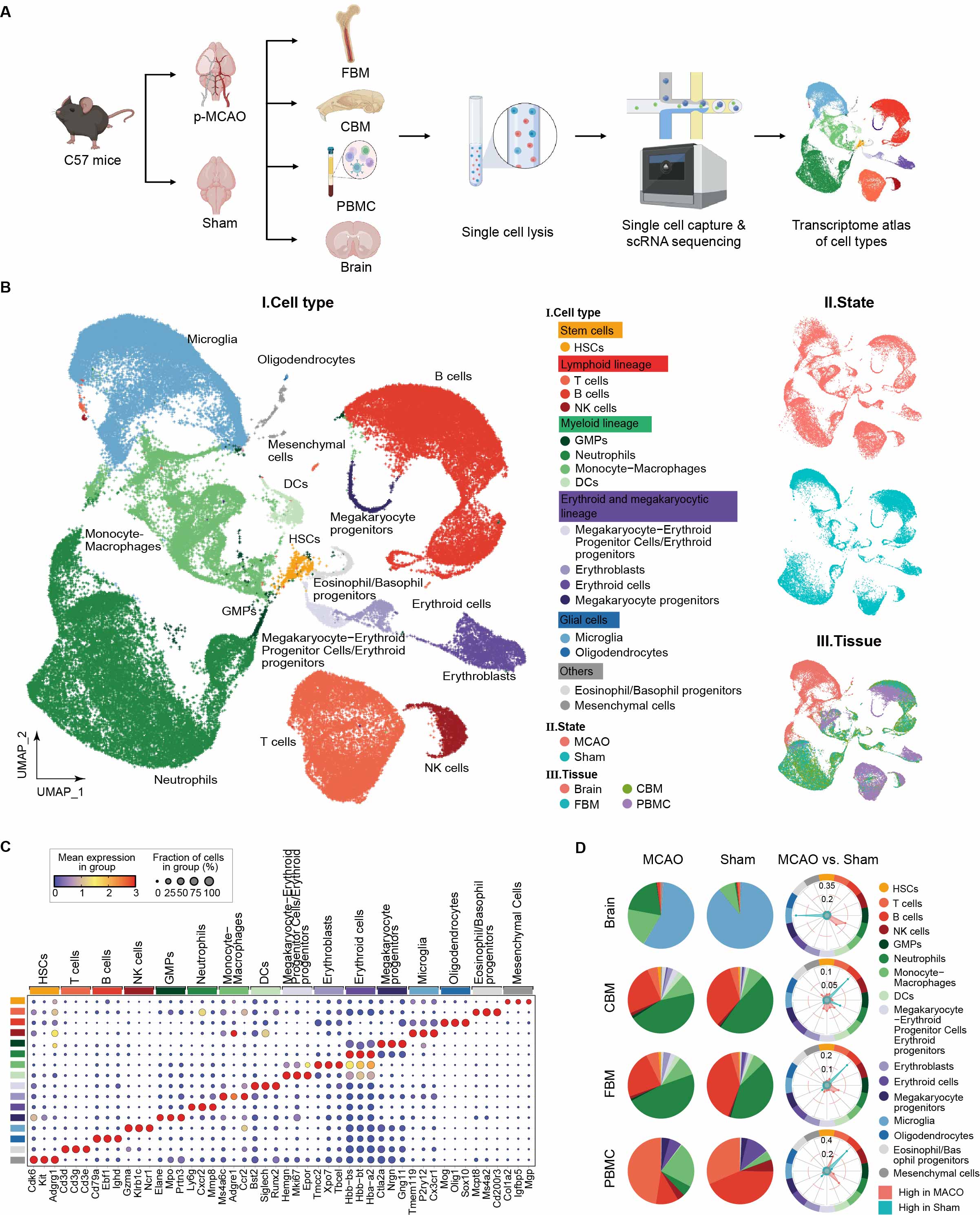
Mouse Stroke
The activation and infiltration of immune cells are hallmarks of ischemic stroke. However, the precise origins and the molecular alterations of these infiltrating cells post-stroke remain poorly characterized. Here, a murine model of stroke (permanent middle cerebral artery occlusion [p-MCAO]) is utilized to profile single-cell transcriptomes of immune cells in the brain and their potential origins, including the calvarial bone marrow (CBM), femur bone marrow (FBM), and peripheral blood mononuclear cells (PBMCs). This analysis reveals transcriptomically distinct populations of cerebral myeloid cells and brain-resident immune cells after stroke. These include a novel CD14+ neutrophil subpopulation that transcriptomically resembles CBM neutrophils. Moreover, the sequential activation of transcription factor regulatory networks in neutrophils during stroke progression is delineated, many of which are unique to the CD14+ population and underlie their acquisition of chemotaxis and granule release capacities. Two distinct origins of post-stroke disease-related immune cell subtypes are also identified: disease inflammatory macrophages, likely deriving from circulating monocytes in the skull, and transcriptionally immature disease-associated microglia, possibly arising from pre-existing homeostatic microglia. Together, a comprehensive molecular survey of post-stroke immune responses is performed, encompassing both local and distant bone marrow sites and peripheral blood.